Protein Structural Features
Welcome to the pre-lab reading for C4: Protein Structural Features. Proteins are the workhorses of the cell, performing a vast array of functions. Their specific function is intimately linked to their three-dimensional structure. Understanding these structures is fundamental to protein modeling.
This guide will walk you through the different levels of protein structure, the forces that stabilize them, and common structural motifs.
1. Levels of Protein Structure
Proteins are complex macromolecules with structures that can be described at four distinct levels: primary, secondary, tertiary, and quaternary. [22]
1.1 Primary (1°) Structure
The primary structure of a protein is its unique sequence of amino acids. [1, 17] Amino acids are linked by covalent bonds called peptide bonds, formed between the carboxyl group of one amino acid and the amino group of the next. [12, 19] This sequence is determined by the gene encoding the protein. [1] Even a small change in the primary structure can alter the protein’s overall shape and function, as seen in diseases like sickle cell anemia. [1]
 Figure 1: Schematic of a protein’s primary structure, showing amino acids linked by peptide bonds. (Source: Wikimedia Commons)
Figure 1: Schematic of a protein’s primary structure, showing amino acids linked by peptide bonds. (Source: Wikimedia Commons)
1.2 Secondary (2°) Structure
Secondary structure refers to regularly repeating local structures stabilized by hydrogen bonds between the carbonyl oxygen and amide hydrogen atoms within the polypeptide backbone. [1, 9] The two most common types of secondary structures are the α-helix and the β-pleated sheet. [9, 16]
α-Helix
An α-helix is a right-handed coiled or spiral conformation. [9, 16] Hydrogen bonds form between every fourth amino acid, specifically between the C=O of one amino acid and the N-H of the amino acid four residues earlier in the sequence. [1, 2] The R groups of the amino acids project outwards from the helix. [1]
 Figure 2: Ribbon diagram of an α-helix. Hydrogen bonds (not shown) between backbone atoms stabilize this structure. (Source: Wikimedia Commons)
Figure 2: Ribbon diagram of an α-helix. Hydrogen bonds (not shown) between backbone atoms stabilize this structure. (Source: Wikimedia Commons)
β-Pleated Sheet
β-pleated sheets are formed by two or more segments of a polypeptide chain (β-strands) lying side-by-side, connected by hydrogen bonds between parts of the two parallel or anti-parallel polypeptide backbones. [1, 11] The R groups extend above and below the plane of the sheet. [1]
- Parallel β-sheets: Strands run in the same N-terminus to C-terminus direction.
- Anti-parallel β-sheets: Strands run in opposite directions.
 Figure 3: Ribbon diagram of a β-pleated sheet (anti-parallel). Hydrogen bonds (not shown) connect adjacent strands. (Source: Wikimedia Commons)
Figure 3: Ribbon diagram of a β-pleated sheet (anti-parallel). Hydrogen bonds (not shown) connect adjacent strands. (Source: Wikimedia Commons)
Turns and Loops
Turns and loops are non-regular secondary structures that connect α-helices and β-sheets. They are often found on the protein surface and can play important roles in protein function and interactions.
Ramachandran Plot
The conformation of the polypeptide backbone is constrained by the dihedral angles phi (φ) and psi (ψ) around the Cα-N and Cα-C bonds, respectively. A Ramachandran plot visualizes the sterically allowed combinations of these angles for each amino acid in a protein. [23, 26, 34] Different secondary structures occupy distinct regions of this plot.
 Figure 4: A Ramachandran plot showing allowed (colored) and disallowed (white) regions for φ and ψ angles. Glycine, with its small side chain, has more allowed conformations. (Source: Wikimedia Commons)
Figure 4: A Ramachandran plot showing allowed (colored) and disallowed (white) regions for φ and ψ angles. Glycine, with its small side chain, has more allowed conformations. (Source: Wikimedia Commons)
1.3 Tertiary (3°) Structure
Tertiary structure refers to the overall three-dimensional shape of a single polypeptide chain, resulting from the folding and packing of its secondary structural elements. [1, 39] This level of structure is stabilized by various interactions between the R groups (side chains) of the amino acids. [1, 4] These interactions include:
- Hydrophobic interactions: Nonpolar side chains tend to cluster in the protein’s interior, away from water. [4, 30]
- Hydrogen bonds: Between polar R groups, and between R groups and the backbone. [4]
- Ionic bonds (salt bridges): Between oppositely charged R groups. [2, 4]
- Disulfide bonds (bridges): Covalent bonds formed between the sulfur atoms of two cysteine residues. These are strong bonds that significantly stabilize the structure. [3, 4]
- Van der Waals forces: Weak, short-range attractive forces between all atoms. [3]
 Figure 5: Example of a protein’s tertiary structure, showing the folding of α-helices and β-sheets into a compact globular shape. (Source: Wikimedia Commons)
Figure 5: Example of a protein’s tertiary structure, showing the folding of α-helices and β-sheets into a compact globular shape. (Source: Wikimedia Commons)
Video explaining the forces stabilizing tertiary structure.
Protein Domains and Motifs
Within the tertiary structure, you can often identify:
- Domains: Distinct, stable, independently folding functional and/or structural units within a protein. [25, 38] Proteins can be composed of one or several domains. Domains often have a specific function, like binding a ligand or catalyzing a reaction. [35]
- Motifs (Supersecondary structures): Recognizable folding patterns involving two or more elements of secondary structure and their connections. [25, 27] Examples include helix-turn-helix, beta-alpha-beta unit, or zinc finger. Motifs can be part of a larger domain. [31]
1.4 Quaternary (4°) Structure
Quaternary structure arises when a protein is made up of two or more polypeptide chains (subunits). [1, 6] It describes the arrangement and interactions of these subunits to form a functional protein complex. [10] The same types of non-covalent interactions and disulfide bonds that stabilize tertiary structure also hold subunits together. [1] Not all proteins have a quaternary structure; some function as single polypeptide chains. [6]
 Figure 6: Hemoglobin is an example of a protein with quaternary structure. It is composed of four polypeptide subunits (two α-chains and two β-chains), each binding a heme group (shown in red/green). [10, 15, 24] (Source: Wikimedia Commons, PDB ID: 1GZX)
Figure 6: Hemoglobin is an example of a protein with quaternary structure. It is composed of four polypeptide subunits (two α-chains and two β-chains), each binding a heme group (shown in red/green). [10, 15, 24] (Source: Wikimedia Commons, PDB ID: 1GZX)
2. Hierarchy of Protein Structure
Understanding the levels of protein structure is like understanding building blocks. The sequence of amino acids (primary) dictates how local regions fold (secondary), which then determines the overall 3D shape of a single chain (tertiary), and finally how multiple chains might assemble (quaternary).
Video from Khan Academy explaining the four levels of protein structure. [44]
3. Factors Determining Protein Structure
The intricate 3D structure of a protein is ultimately determined by its amino acid sequence (primary structure). [17] However, the process of folding into its functional, native conformation can be influenced by:
- The cellular environment: pH, temperature, salt concentration.
- Chaperone proteins: These assist in the correct folding of other proteins, preventing misfolding and aggregation. [14]
Changes in the environment, such as extreme temperature or pH, can disrupt the weak bonds maintaining secondary, tertiary, and quaternary structures, leading to denaturation, where the protein loses its shape and function. [1]
4. Visualizing Protein Structures
Understanding protein structure is greatly aided by visualization tools. Experimental techniques like X-ray crystallography, NMR spectroscopy, and cryo-electron microscopy provide atomic coordinates for protein structures. These coordinates are stored in public databases.
The Protein Data Bank (PDB)
The Protein Data Bank (PDB) is the primary global repository for 3D structural data of large biological molecules, including proteins and nucleic acids. [8, 13, 36] Each structure in the PDB is assigned a unique 4-character PDB ID. [21]
 Figure 7: Logo of the RCSB Protein Data Bank, a key resource for protein structures. (Source: RCSB PDB)
Figure 7: Logo of the RCSB Protein Data Bank, a key resource for protein structures. (Source: RCSB PDB)
Molecular Visualization Software
Various software programs allow researchers to visualize, analyze, and manipulate these 3D structures. Common examples include PyMOL, ChimeraX, and Mol*. [18, 37] These tools can render proteins in different representations (e.g., ribbons, spheres, surfaces) to highlight various features. [33, 42]
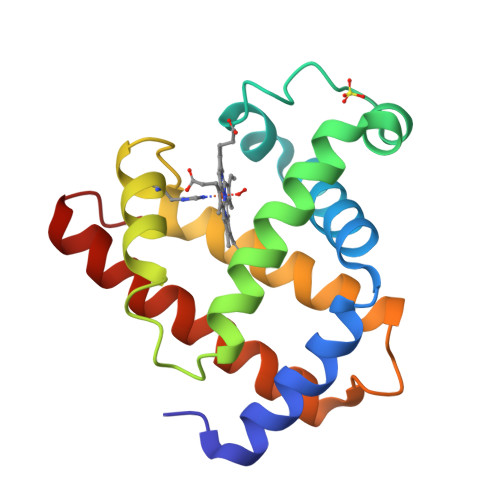
Below is an example of a protein structure (Myoglobin, PDB ID: 1MBO) visualized using Mol* from the RCSB PDB. Click on the hotspots to learn about different structural features.
5. Conclusion
The structure of a protein is paramount to its function. From the linear sequence of amino acids to the complex assembly of multiple subunits, each level of organization contributes to the protein’s final architecture and biological role. In this course, you will learn to model these structures, predict their properties, and understand how they interact with other molecules.
This pre-lab reading provides the foundational knowledge you’ll need for our upcoming lab sessions on protein modeling. Make sure you are comfortable with these concepts.
- Resources
- API
- Sponsorships
- Open Source
- Company
- xOperon.com
- Our team
- Careers
- 2025 xOperon.com
- Privacy Policy
- Terms of Use
- Report Issues